Connexin Field Guide
Connexinplexity
Connexins are a large gene family, and we suspect that all cells in vertebrates use them either for making gap junctions or for paracrine/autocrine signaling as hemichannels. This family has been challenging to understand due to nomenclature issues and the complexity of members (20 in humans, 41 in zebrafish). Recent work from Mikalsen et al. clarified the gene family relationships and we sought to connect zebrafish and human genes via expression, mutant phenotypes, evolutionary relationships, etc. See Lukowicz RL, et al.; bioRxiv. To help the community understand ‘Connexinplexity’, and serve as a platform for human disease models, we provide a number of resources below.
A Field Guide with Human/Zebrafish Connexin genes, proteins, papers, tools.
A simple list of names/IDs for zebrafish connexins.
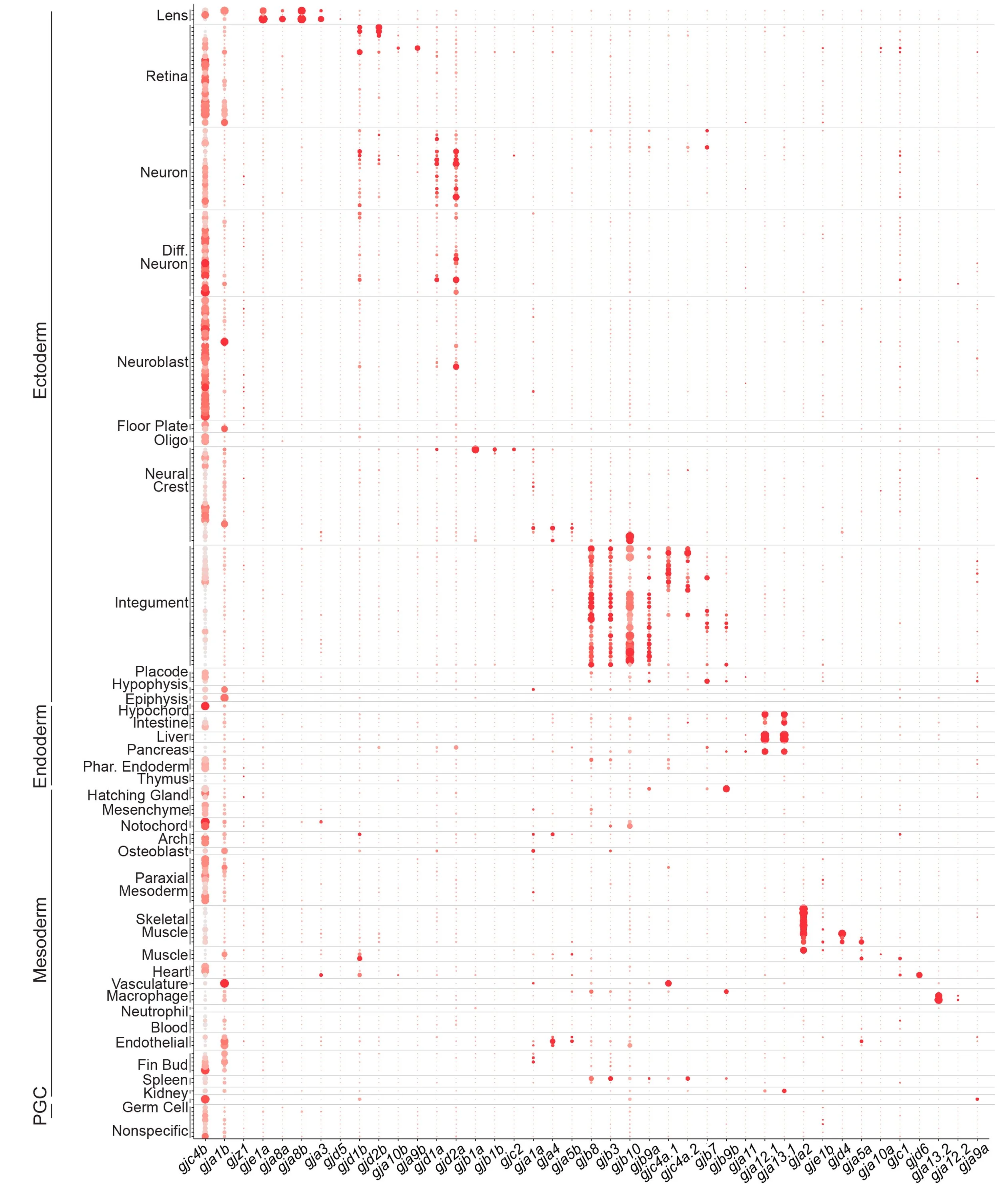
Overview of connexin expression (Fig.1), details (Fig.2), integument (Fig.3) (see paper for info).
Atlas 1.0 - Connexinplexity
The field guide and paper leverage our scRNA-seq atlas, but this effort required an update of the zebrafish gene model (we modified the GTF from Lawson et al.) and a reanalysis of the Atlas 1.0 data. Below are useful tools and data from the work.
UCSC Cell Browser, Updated GTF, Clusters (TableS3), Seurat v3.1.5 (Connexinplexity.rds, Connexinplexity.rmd, code).
Atlas
Atlas of zebrafish development at single cell resolution
single-cell RNA-seq technologies are rapidly increasing our understanding of organismal biology and provides new opportunities to understand the systems biology of development. We, along with Chuck Kimmel, John Postlethwait, and Monte Westerfield, have initiated a project to map zebrafish at single cell resolution during development and at adulthood (NIH R24OD026591). Links to our evolving analyses and publications can be found below.
Atlas 1.0
Our initial efforts have been put together in the manuscript Farnsworth DR, et al.; bioRxiv and DevBio (and see here for more pubs). Useful data from that paper are hosted here:
UCSC Cell Browser, Clusters (TableS1, TableS2), Seurat v2 (Atlas.rds, Atlas.rmd, code).
The fastq files from all experiments are available at the NCBI SRA, accession: PRJNA564810.
CRISPR
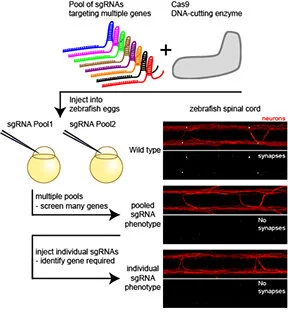
CRISPRscreen
CRISPR allows for unprecedented control over modifying genomes. We took advantage of CRISPR's high rate of mutagenesis to figure out how to perform in vivo reverse genetic screens in a zebrafish (Paper). We screened for electrical synapse defects but also found that the approach was applicable too many different developmental processes. We also wrote a methods chapter to help out with the approach (Method). At the time we were using Cas9 RNA, but it is very clear that Cas9 protein is much more effective. We recommend following the excellent advise of Alexa Burger from the Mosimann group as this has worked with extremely high mutagenesis efficiency for us (Cas9 protein).
RNAmapper
RNAmapper
One of the main goals in lab is to be able to quickly interrogate gene function in vivo in a vertebrate system. As zebrafish geneticists we love to be able to make mutations in genes and then assess the phenotypic outcome. However, classic forward genetic approaches, in which random mutations are generated in the genome, are notoriously slow at the stage of identifying the mutated gene. It’s akin to finding a needle in a haystack. To speed the process we developed a next generation sequencing method to quickly map and identify mutated genes. We generated code and a Galaxy implementation to help the community map mutations. Learn more about RNAmapper.
We co-published our work with along with Jonathan Hill who was working in the Yost lab at the time. He generated a related pipeline, and we encourage you to try it out in parallel to ours (MMAPR).